Green Fluorescent Protein (GFP) engineering and screening of genome-wide regulatory element libraries in E. coli
Protein engineering has many applications, from antibody and enzyme engineering to studying the sequence-function relationships and structural characteristics. Whether the end goal is functional genomics research or industrial applications, it requires being able to generate defined, trackable, stable genomic edits, followed by functional screening of resulting variants. Traditional methods – like site-directed or random mutagenesis of plasmid-expressed genes – fall short when it comes to providing efficient and cost-effective ways of generating large numbers of engineered variants to probe a significant portion of the protein sequence space.
Being able to rapidly and efficiently engineer heterologous proteins requires that genetic edits are generated in a highly-multiplexed manner. To demonstrate how this can be done with the Onyx platform, we designed an edit library for a green fluorescent protein (GFP) integrated into the E. coli MG1655 genome. We designed 723 specific edits targeting known mutational hotspots – residues previously implicated in improving protein stability or altering its activity. The entire library was constructed in one Onyx run. With a few hundred new variants on hand, we went straight to phenotyping.
Because the GFP was engineered directly on the chromosome, using a single gene copy under the expression of a constitutive promoter, we could easily detect and quantify the changes in fluorescence. Screening of the library variants quickly revealed several edits that enhanced fluorescence. Additionally, we performed screenings using excitation-emission wavelengths for the yellow and blue fluorescent proteins, along with spectral scans of selected variants. Phenotyping revealed that certain edits altered fluorescence intensity while others caused yellow or blue shifts in emission spectra. The hits included previously known and novel sequence variants.
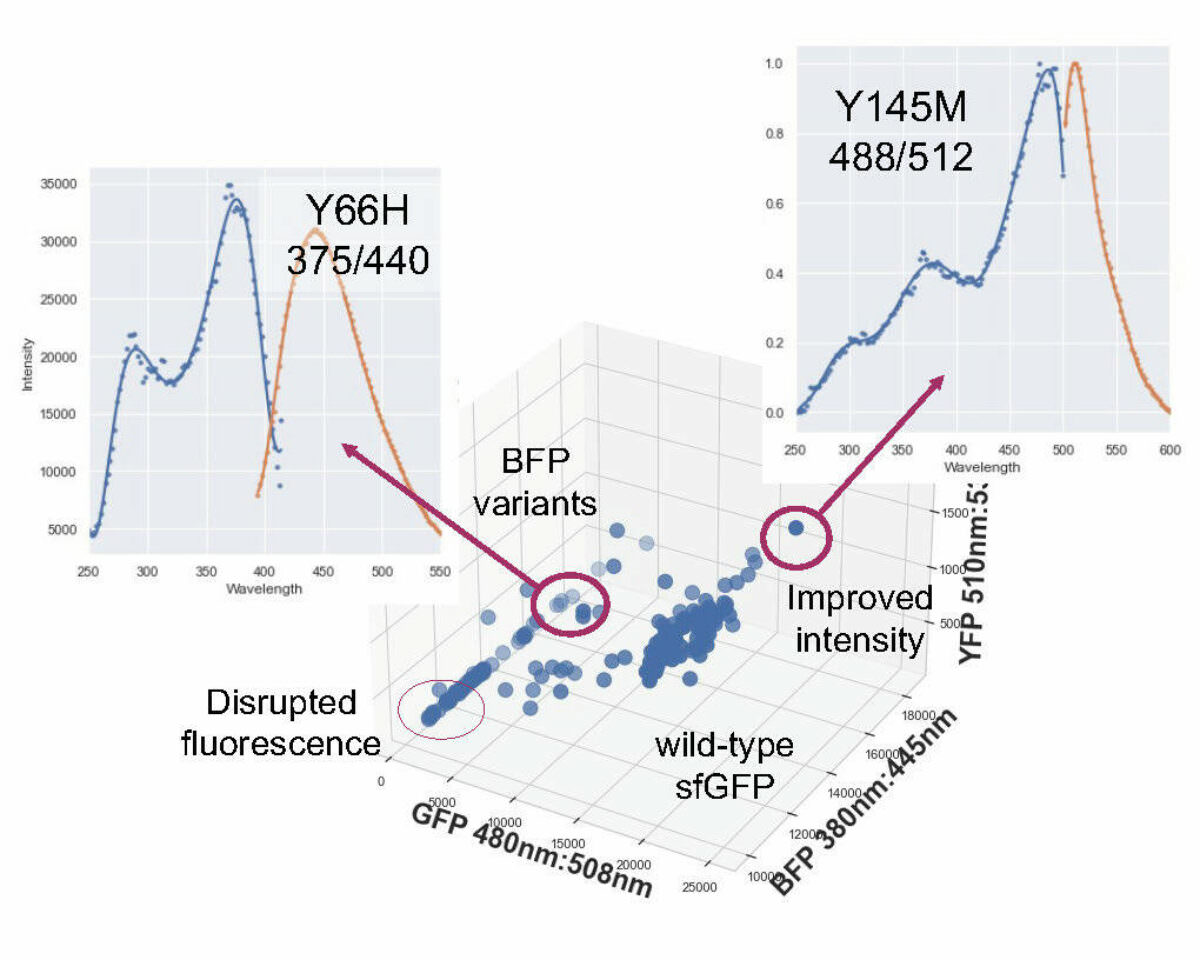
Fluorescence profile of the screened GFP library showing variants with improved fluorescence intensity and spectral shifts.
We then asked ourselves: what else could we do with a genomically integrated GFP? How about testing a library of 8,284 native regulatory elements and quantifying their strength. We extracted sequences of all hypothetical promoter and ribosomal binding sites (RBS) from the E. coli genome and inserted those in front of GFP, enabling us to sort thousands of native regulatory sequences according to their activity and function.
Generate any library at the push of a button.
The Onyx gives you the flexibility and scalability to develop virtually limitless libraries. Create insertions, deletions, substitutions, or full saturation mutagenesis, in a single gene, a pathway, or the whole genome.
- Copyright © 2025 Inscripta, Inc. All rights reserved.
- Legal Notices
- Privacy Policy
