Powerful support for scientists.
Videos
Presentations & webinars
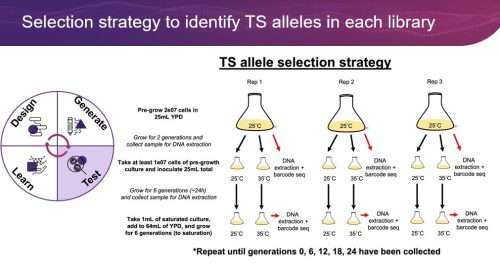
Temperature sensitive (TS) alleles are mutations in essential genes that allow for growth at permissive temperatures and restrict or reduce growth at non-permissive temperatures. These conditional mutations are useful in academic settings for basic study of essential genes, and they are useful in bio-industrial settings as many essential genes are central to the production of high-value compounds. While critical for the study and use of essential genes, TS alleles are notoriously difficult to find. During this webinar, we will showcase how we leverage the precise, trackable genome editing of the Onyx Platform to discover novel TS alleles in yeast and E. coli. Following genome engineering, selection experiments have led to the identification of many potential TS alleles. Initial validation experiments are proving that many of these potential hits are novel TS alleles that show distinct growth properties when comparing permissive and non-permissive growth temperatures. These novel conditional mutants will function as invaluable tools for future studies and industrial applications of essential genes. This work highlights the effectiveness of Inscripta’s Digital Genome Engineering platform for novel discoveries in both yeast and E. coli.
Manufacturing of proteins, mRNA or small molecules requires comprehensive strain engineering approaches to increase yield, productivity, and robustness of the strain. While some of these goals can be accomplished with standard genetic engineering methods, others require a whole-genome, exploratory approach that can take many months. With the Onyx® Digital Genome Engineering platform, you can go after each of these objectives simultaneously by generating diverse strain libraries in a simple, automated workflow. Insertions, deletions, and substitutions across multiple targets, such as promoters, coding regions or global regulators, can be rapidly introduced to improve both specific functions and the overall manufacturability of the strain. In this webinar we will demonstrate how this approach helped improve production of heterologous proteins in E. coli and S. cerevisiae.
E. coli B‑strains, such as BL21, and their derivatives are often used for protein expression due to their optimized amino acid production, ATP utilization, lon and ompT protease deficiency, and reduced acetate accumulation. However, optimizing B‑strains for increased protein production using modern approaches to genome engineering has not been reported. Here we apply CRISPR-based, high-throughput genome-wide and pathway-specific editing to improve the expression of an enzyme. With shallow sampling of the genome-edited libraries, we identify variants with greater than 2‑fold improvement in active protein expression. This work demonstrates how rapid genome engineering can be applied to significantly enhance protein expression in E. coli.
The ability to move rapidly through the Design-Generate-Test-Learn (DGTL) cycle is essential for effective forward engineering of microbial phenotypes. The Onyx® platform is an automated CRISPR-based genome editing technology that enables rapid diversity generation. To optimize the DGTL cycle, effective diversity generation must be paired with an appropriate screening strategy for isolated genetic variants. Here we discuss the benefits of creating a higher edit diversity in cell populations while using a shallow screening approach with high-throughput and automated phenotyping systems such as Rapid Fire Mass Spec and plate-based fluorometric assays. We demonstrate the effectiveness of this approach using both metabolic and strain engineering examples, including a 14,000-fold improvement in lysine biosynthesis in E. coli and 4‑fold increase in heterologous protein production in S. cerevisiae.
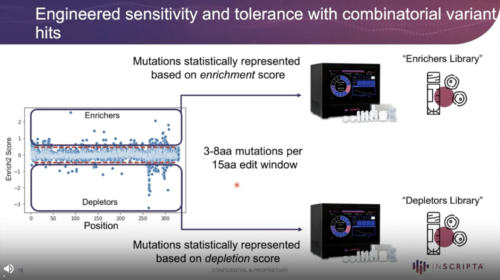
Members of the cinnamic acid and stilbenoid chemical families are common intermediates in the production of important bioindustrial compounds. However, these molecules can inhibit microbial growth and stifle production capabilities. Since the underlying mechanism of action is unclear, ways to ameliorate inhibition can be time consuming and difficult to identify. Here we use automated, high-throughput, precise and trackable genome editing to probe biological responses to p‑coumaric acid and resveratrol. Using a combination of genome-wide and gene-specific edits, we identify genes and individual residues that show enrichment or depletion in the presence of p‑coumaric acid or resveratrol. This method represents a new approach to small molecule mechanisms-of-action discovery, providing target leads that can subsequently be used to engineer tolerance or enhance sensitivity to antimicrobial compounds.
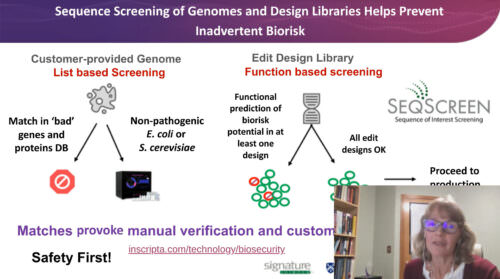
Bioengineering technologies hold enormous potential to solve many of our world’s pressing problems. However, they also pose biosecurity considerations. Inscripta® is committed to safe use of its genome engineering technology and has implemented multi-faceted biosecurity system designed to identify and help prevent biorisk potential of engineered organisms. Drs. Ternus, Treangen, and Vitalis will describe approaches and challenges of genome engineering biosecurity and highlight a computational tool that can predict protein function of large numbers of engineered sequence variants. The SeqScreen platform was developed by a multi-institutional team led by Signature Science and Rice University under the IARPA Fun GCAT program. Case studies will illustrate how SeqScreen serves as a comprehensive tool to report features and predicted function of novel sequences arising from bioengineering.
This is an ideal forum for objective first-hand updates on both marketed and emerging products across the sector with more than 375 public and private companies across the entire health care spectrum. Cowen research analysts will host public companies in fireside chat sessions and corporate panel discussions to engage in enlightening discussion with management teams.
Join the Synthetic Biology Enabling Technologies Panel on March 7 at 10:30 am ET
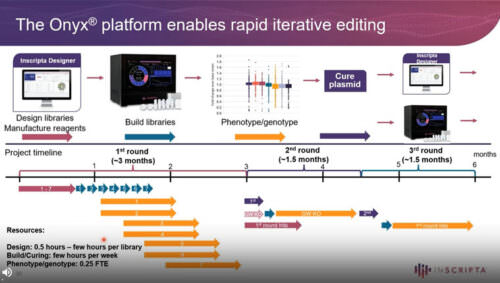
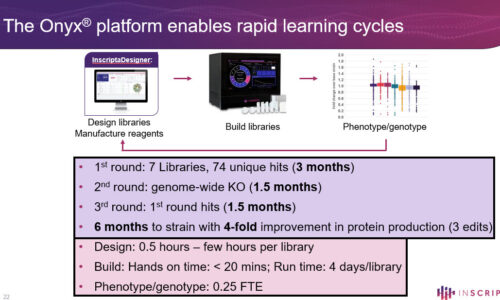
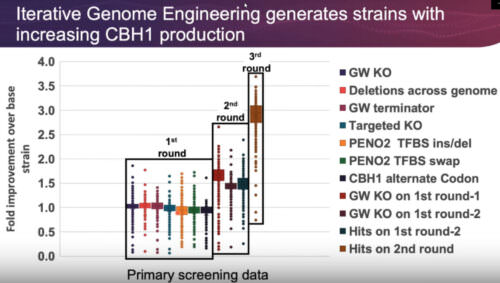
The Design – Generate – Test – Learn (DGTL) cycle represents an efficient approach to biological engineering, from single-gene to whole-genome scale. Significant improvement can be achieved in a short amount of time by recombining beneficial edits through iterative cycles of genome editing. In order to cut down development times, all aspects of the DGLT cycle must be optimized. The Onyx™ Digital Genome Engineering Platform automates the Design and Generate steps, allowing the user to create thousands of edits, identify the beneficial variants, and construct new libraries based on the improved strains – all in an integrated, seamless workflow. Here we demonstrate how Iterative Genome Engineering can be used to discover novel protein variants, optimize expression, and improve heterologous protein production in yeast and E. coli strains.
Synthetic biology is disrupting diverse markets from chemicals to food, cosmetics, materials, and therapeutics. Whether you’re a metabolic engineer optimizing lysine biosynthesis in E. coli or seeking to conduct a genome-wide functional genomics screen in yeast, Digital Genome Engineering is the key to unlocking the limitless applications of synthetic biology. Realizing the full potential of this game changing technology in your research requires accessing the latest tools, understanding industry trends, and learning from best practices of industry leaders.
In partnership with SynBioBeta, Inscripta® is bringing together synthetic biology thought leaders for an innovative, FREE 2‑hour symposium. The sessions will feature perspectives from industry leaders, highlight the latest innovations in Digital Genome Engineering, and provide a vision for the future of SynBio.
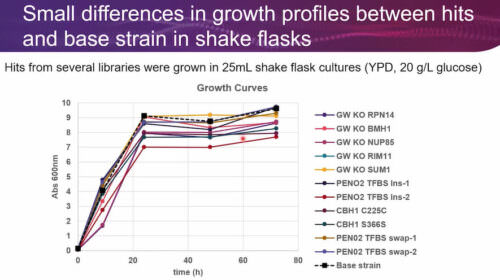
Heterologous protein production is an indispensable tool in biotechnology and biopharma for manufacturing enzymes, protein therapeutics, and more. Generating robust production strains involves strategies that target both the protein expression cassette and the host genome. However, despite advances in strain engineering, optimization of protein production is still constrained by low-throughput and laborious methods that limit the success of strain optimization efforts.
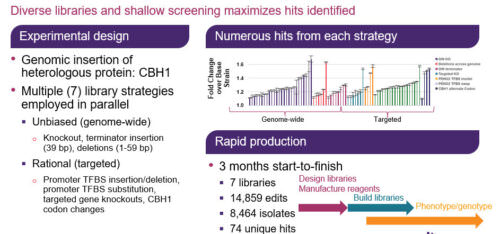
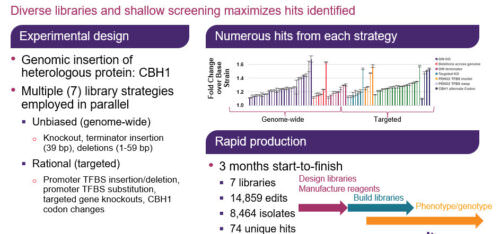
In this GEN webinar, our distinguished presenter, Dr. Eric Abbate, will demonstrate a novel multiplexed genome-wide editing approach to optimize the cellobiohydrolase I enzyme (CBH1) production in S. cerevisiae. Additionally, we will learn how multiplexed and trackable editing libraries enable the discovery of new targets. These libraries comprise a wide range of edit types and targets, including genome-wide and targeted knockouts, short deletions, alternate codon, transcription factor binding sites, and terminator libraries. Finally, Dr. Abbate will describe how to identify beneficial edits across targets involved in transcription, translation, glycosylation, secretion, protein degradation, and other cellular processes, as well as evaluate the success of using targeted vs. untargeted libraries for strain engineering.

— Tycho Peterson, Managing Director, Life Science Tools & Diagnostics, J.P. Morgan
— Ipsita Smolinski, Managing Director, Capitol Street
Abstract: Join the panel to discuss the genetic technology known as synthetic biology. Two industry heavyweights — Ginkgo & Inscripta — will describe genomic engineering & organism technologies. The White House will weigh in on how it evaluates technologies and partnerships to further US science and technology priorities. One key trade association will discuss the impact synthetic biology has on non-health applications, fragrances and other flavors.

CRISPR editing has revolutionized plant breeding, and the MAD7 CRISPR nuclease developed by Inscripta<sup>TM</sup> is democratizing access to CRISPR technologies. Hudson River Biotechnology has developed a proprietary, validated molecular breeding workflow called MAD TiGER, which stands for Target identification, Guide selection, Entry into the cell and Regeneration using the MAD7 nuclease. MAD TiGER enables molecular breeding in an end-to-end fashion, providing innovative solutions to the typical barriers encountered in CRISPR plant breeding projects, such as cell entry and regeneration. The MAD TiGER workflow uses the MAD7 nuclease, nanotechnology and various synthetic gels to enable faster onset of cell divisions in regenerating and de-differentiating protoplasts towards micro-calli formation. During this webinar, Gabino Sanchez of Hudson River Biotechnology will provide an overview of the MAD TiGER workflow and discuss examples of using an integrated workflow for molecular breeding. You will also learn more about the MAD7 nuclease from Inscripta.
Inscripta’s Senior Director of Cell Biology, Patrick Westfall, led a panel discussion “Biological Engineering the Next Generation of Materials and Chemicals” with Kirsten Benjamin, Vice President of R&D Amyris, and Andrew Horwitz of Sestina Bio. Westfall asked the panelists about the process of selecting products they want to make and what considerations go into making those decisions.
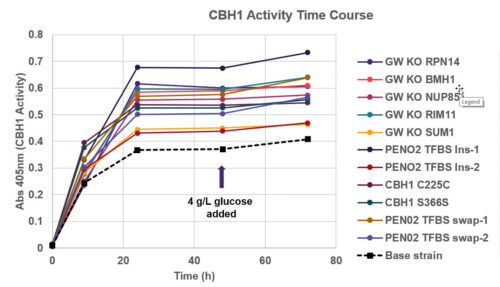
Heterologous protein production is an indispensable tool in biotechnology and biopharma for manufacturing enzymes, protein therapeutics, and more. Generating robust production strains involves strategies that target the protein itself as well as the host genome. Despite advances in strain engineering, optimization of protein production is still limited by the ability to access the broad sequence space, constrained by traditional, low-throughput, and laborious methods like promoter substitutions or random mutagenesis. Here we use a genome-wide, multiplexed, targeted editing approach that can generate diverse edit types in both the heterologous gene and across the host genome to optimize production of cellobiohydrolase I enzyme (CHB1) in S. cerevisiae. The edits conferring improved CBH1 expression span cellular processes important for protein production, including transcription, translation, secretion, and protein degradation pathways.
Discovery in biological research was historically driven by empirical methods. The advent of biological engineering, propelled by technological developments and new tools, has made rational hypothesis-driven design and engineering of biological systems more feasible and common. Although they are often seen as opposing approaches, the empirical and rational methods can work together to accelerate fundamental research and product development. Whereas forward engineering is guided by informed design of known biological pathways and processes, the exploratory empirical methods allow to expand the design space beyond the scope of current knowledge. This webinar discusses the advantages of each method and how they could be implemented in tandem to advance biological engineering.
Heterologous protein expression in model organisms has many applications, including protein engineering, production of industrial enzymes and therapeutics, and structure-function discovery. Heterologous proteins are commonly expressed from plasmids to facilitate cloning and genetic manipulation. However, plasmid-based expression has many drawbacks, such as reduced stability and limited control over gene copy number. With CRISPR editing, chromosomal gene integration and subsequent editing has become more accessible, enabling thorough examination of protein expression and function. In this webinar, we demonstrate how we generated an
Biological responses to environmental stress can arise from multiple genetic solutions encoding tolerance mechanisms. In order to investigate the response to osmotic pressure in E. coli, we used an automated CRISPR editing workflow to generate a diverse cell population that included gene knockouts and different strength promoter substitution libraries targeting almost every gene in the genome. The resulting 25,000-variant pooled library was studied under a titrated salt challenge and the population dynamics were tracked using plasmid barcodes. By mapping 300,000 data points, we identified clear enrichment and depletion patterns and connected those back to functional genome annotations. We were able to rapidly validate our experiment with the performance of known salt stress tolerance loci, such as rpoE and osmE, and discover new gene targets.
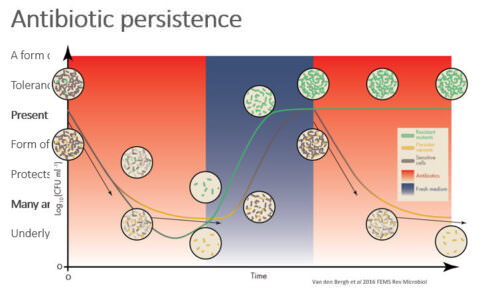
Bacterial persistence is a potential cause of antibiotic therapy failure. Recent studies have shown that persistence is a highly evolvable trait, reaching varying frequencies within populations, depending on the selective conditions that are applied (e.g. the treatment frequency). Theory predicts that population bottlenecks, frequently encountered during host-to-host transmission and antibiotic treatment, have far-reaching effects on the evolutionary dynamics of persistence. Here, we used a combination of experimental evolution and barcoded knockout libraries to examine this hypothesis. Small bottlenecks were found to restrict the adaptive potential of populations and result in more heterogeneous evolutionary outcomes, suggest that the fitness landscape of persistence has a rugged topography. Furthermore, sequencing data revealed genes that are potentially involved in persistence, including previously known as well as novel targets.
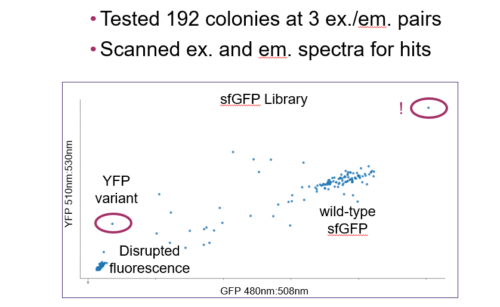
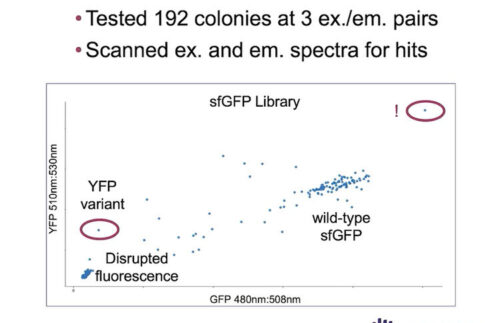
During this webinar, discover how Inscripta experts generated an Escherichia coli saturation mutagenesis library against an integrated green fluorescent protein (GFP) using CRISPR-based technology, in a few short weeks.

The panel of industry leaders discuss technologies and microbial applications of synthetic biology in biopharma bioproduction.
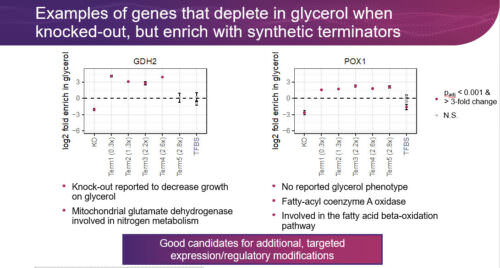
In this webinar, we will discuss our work using the Onyx platform to perform genome-wide engineering of S. cerevisiae for various applications including target discovery to increase glycerol utilization, strain optimization, and forward engineering. These applications are enabled by Onyx’s ability to deliver diverse edit types that regulate gene function and expression beyond simple gene knock-outs. In total, dozens of libraries of engineered cells, containing over 100,000 precise genomic edits have been created and tested to accelerate forward engineering and genomic discovery.
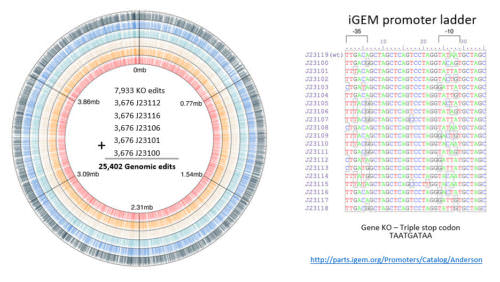
Lysine, predominately used as an animal feed supplement, is a multi-billion-dollar industry however supply is limited due to the cost of current production methods. This webinar will share case studies how Inscripta leveraged its new CRISPR-based technology to design, generate, and screen libraries totaling > 200,000 single edits to improve lysine production in E. coli. Dozens of beneficial edits across the entire genome were discovered to accelerate the Design-Generate-Test-Learn cycle and increase the lysine production by 14,000-fold — ushering in the next era of genome editing.
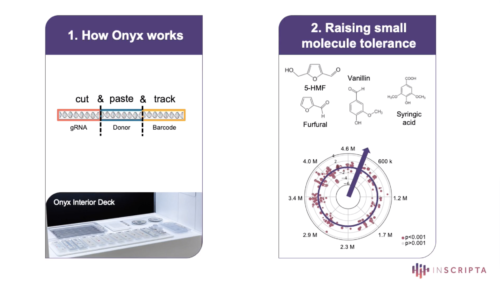
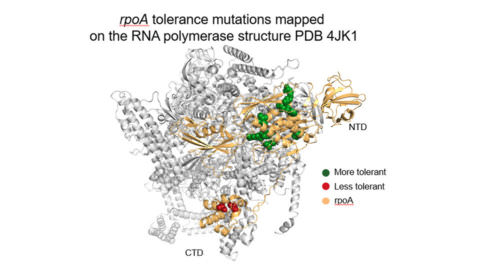
Since its introduction, CRISPR has become a core genome editing tool that has transformed biological research. First-generation CRISPR technologies were limited in scalability, accessibility, edit variety, and ease of use, restricting their potential. Inscripta’s Onyx™ platform addresses these limitations by combining easy-to-use, intuitive software with a push-button automated benchtop device, enabling high-efficiency, massively-parallel, precision-engineered edits to Saccharomyces cerevisiae and Escherichia coli genomes. In this webinar , Tyson Shepherd will present applications in E. coli that leverage this platform. He will show how a library totaling more than 25,000 different designs and a separate library of 900 designs yielded new biochemical insights underpinning tolerance to a panel of growth-inhibitory compounds. These applications demonstrate the power of the Onyx platform to usher in a new era of genome editing.
In this study, we used the high-throughput CRISPR-based Onyx platform to perform saturation mutagenesis on four different essential genes involved in cell envelope synthesis in E. coli. 22,790 edits were designed. We used these saturation mutagenesis libraries to probe the essentiality of all different residues. Edits that could not be introduced suggests those residues are essential for protein function. We identified known essential amino acids (i.e. catalytic residues and residues involved in substrate binding), thereby validating our experimental approach. Several residues that were previously not known to be essential were identified. We expect our results to offer vastly improved insights into protein function, help fight against antibiotic resistance and aid structure-based drug design targeted against these essential proteins.
In this webinar, we discuss our work using the Onyx platform to perform genome-wide engineering of S. cerevisiae for various applications including target discovery to increase glycerol utilization, strain optimization, and forward engineering. These applications are enabled by Onyx’s ability to deliver diverse edit types that regulate gene function and expression beyond simple gene knock-outs. In total, dozens of libraries of engineered cells, containing over 100,000 precise, genomic edits have been created and tested to accelerate forward engineering and genomic discovery.
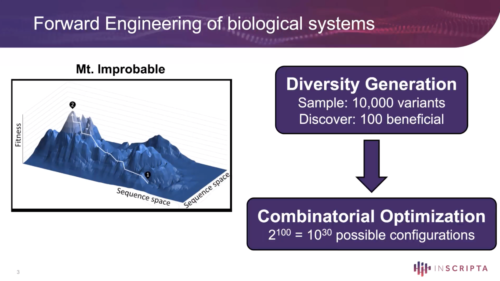
Many of the governing rules of biology are still not known nor well understood. Thus, forward engineering of biological systems will continue to rely on empirical methods. A central challenge in forward engineering is to find genetic interventions that lead to improved function. Combinatorial optimization (fueled by large-scale diversity generation) takes ideas that show promise and recombines them in novel configurations to leverage the principles of evolutionary optimization. This optimization process is further accelerated through strategies that incorporate machine learning. Such models are interrogated to efficiently guide new library designs. Strategies and tools to effectively apply the principles of diversity generation (and combinatorial optimization) have been developed over the last two decades and are now routinely used to rapidly engineer biological systems.
Significant potential exists to harness the power of biology to better humanity and the environment, however, substantial challenges remain. Forward engineering of biological systems will continue to rely on empirical methods. Diversity generation is one of two key principles needed to reach desired outcomes in a rapid, efficient and cost-effective manner. Sequence space is vast. A central challenge in forward engineering is to find genetic interventions that lead to improved function. A large-scale diversity generation approach, where many thousands of ideas are rapidly tested, has proved to be crucial to addressing this challenge. Strategies and tools to effectively apply the principles of diversity generation (and combinatorial optimization) have been developed over the last two decades and are now routinely used to rapidly engineer biological systems.

Temperature sensitive (TS) alleles are mutations in essential genes that allow for growth at permissive temperatures and restrict or reduce growth at non-permissive temperatures. These conditional mutations are useful in academic settings for basic study of essential genes, and they are useful in bio-industrial settings as many essential genes are central to the production of high-value compounds. While critical for the study and use of essential genes, TS alleles are notoriously difficult to find. During this webinar, we will showcase how we leverage the precise, trackable genome editing of the Onyx Platform to discover novel TS alleles in yeast and E. coli. Following genome engineering, selection experiments have led to the identification of many potential TS alleles. Initial validation experiments are proving that many of these potential hits are novel TS alleles that show distinct growth properties when comparing permissive and non-permissive growth temperatures. These novel conditional mutants will function as invaluable tools for future studies and industrial applications of essential genes. This work highlights the effectiveness of Inscripta’s Digital Genome Engineering platform for novel discoveries in both yeast and E. coli.

Bacterial persistence is a potential cause of antibiotic therapy failure. Recent studies have shown that persistence is a highly evolvable trait, reaching varying frequencies within populations, depending on the selective conditions that are applied (e.g. the treatment frequency). Theory predicts that population bottlenecks, frequently encountered during host-to-host transmission and antibiotic treatment, have far-reaching effects on the evolutionary dynamics of persistence. Here, we used a combination of experimental evolution and barcoded knockout libraries to examine this hypothesis. Small bottlenecks were found to restrict the adaptive potential of populations and result in more heterogeneous evolutionary outcomes, suggest that the fitness landscape of persistence has a rugged topography. Furthermore, sequencing data revealed genes that are potentially involved in persistence, including previously known as well as novel targets.

The panel of industry leaders discuss technologies and microbial applications of synthetic biology in biopharma bioproduction.

In this study, we used the high-throughput CRISPR-based Onyx platform to perform saturation mutagenesis on four different essential genes involved in cell envelope synthesis in E. coli. 22,790 edits were designed. We used these saturation mutagenesis libraries to probe the essentiality of all different residues. Edits that could not be introduced suggests those residues are essential for protein function. We identified known essential amino acids (i.e. catalytic residues and residues involved in substrate binding), thereby validating our experimental approach. Several residues that were previously not known to be essential were identified. We expect our results to offer vastly improved insights into protein function, help fight against antibiotic resistance and aid structure-based drug design targeted against these essential proteins.


Temperature sensitive (TS) alleles are mutations in essential genes that allow for growth at permissive temperatures and restrict or reduce growth at non-permissive temperatures. These conditional mutations are useful in academic settings for basic study of essential genes, and they are useful in bio-industrial settings as many essential genes are central to the production of high-value compounds. While critical for the study and use of essential genes, TS alleles are notoriously difficult to find. During this webinar, we will showcase how we leverage the precise, trackable genome editing of the Onyx Platform to discover novel TS alleles in yeast and E. coli. Following genome engineering, selection experiments have led to the identification of many potential TS alleles. Initial validation experiments are proving that many of these potential hits are novel TS alleles that show distinct growth properties when comparing permissive and non-permissive growth temperatures. These novel conditional mutants will function as invaluable tools for future studies and industrial applications of essential genes. This work highlights the effectiveness of Inscripta’s Digital Genome Engineering platform for novel discoveries in both yeast and E. coli.

Manufacturing of proteins, mRNA or small molecules requires comprehensive strain engineering approaches to increase yield, productivity, and robustness of the strain. While some of these goals can be accomplished with standard genetic engineering methods, others require a whole-genome, exploratory approach that can take many months. With the Onyx® Digital Genome Engineering platform, you can go after each of these objectives simultaneously by generating diverse strain libraries in a simple, automated workflow. Insertions, deletions, and substitutions across multiple targets, such as promoters, coding regions or global regulators, can be rapidly introduced to improve both specific functions and the overall manufacturability of the strain. In this webinar we will demonstrate how this approach helped improve production of heterologous proteins in E. coli and S. cerevisiae.

E. coli B‑strains, such as BL21, and their derivatives are often used for protein expression due to their optimized amino acid production, ATP utilization, lon and ompT protease deficiency, and reduced acetate accumulation. However, optimizing B‑strains for increased protein production using modern approaches to genome engineering has not been reported. Here we apply CRISPR-based, high-throughput genome-wide and pathway-specific editing to improve the expression of an enzyme. With shallow sampling of the genome-edited libraries, we identify variants with greater than 2‑fold improvement in active protein expression. This work demonstrates how rapid genome engineering can be applied to significantly enhance protein expression in E. coli.

The ability to move rapidly through the Design-Generate-Test-Learn (DGTL) cycle is essential for effective forward engineering of microbial phenotypes. The Onyx® platform is an automated CRISPR-based genome editing technology that enables rapid diversity generation. To optimize the DGTL cycle, effective diversity generation must be paired with an appropriate screening strategy for isolated genetic variants. Here we discuss the benefits of creating a higher edit diversity in cell populations while using a shallow screening approach with high-throughput and automated phenotyping systems such as Rapid Fire Mass Spec and plate-based fluorometric assays. We demonstrate the effectiveness of this approach using both metabolic and strain engineering examples, including a 14,000-fold improvement in lysine biosynthesis in E. coli and 4‑fold increase in heterologous protein production in S. cerevisiae.

Members of the cinnamic acid and stilbenoid chemical families are common intermediates in the production of important bioindustrial compounds. However, these molecules can inhibit microbial growth and stifle production capabilities. Since the underlying mechanism of action is unclear, ways to ameliorate inhibition can be time consuming and difficult to identify. Here we use automated, high-throughput, precise and trackable genome editing to probe biological responses to p‑coumaric acid and resveratrol. Using a combination of genome-wide and gene-specific edits, we identify genes and individual residues that show enrichment or depletion in the presence of p‑coumaric acid or resveratrol. This method represents a new approach to small molecule mechanisms-of-action discovery, providing target leads that can subsequently be used to engineer tolerance or enhance sensitivity to antimicrobial compounds.

This is an ideal forum for objective first-hand updates on both marketed and emerging products across the sector with more than 375 public and private companies across the entire health care spectrum. Cowen research analysts will host public companies in fireside chat sessions and corporate panel discussions to engage in enlightening discussion with management teams.
Join the Synthetic Biology Enabling Technologies Panel on March 7 at 10:30 am ET

The Design – Generate – Test – Learn (DGTL) cycle represents an efficient approach to biological engineering, from single-gene to whole-genome scale. Significant improvement can be achieved in a short amount of time by recombining beneficial edits through iterative cycles of genome editing. In order to cut down development times, all aspects of the DGLT cycle must be optimized. The Onyx™ Digital Genome Engineering Platform automates the Design and Generate steps, allowing the user to create thousands of edits, identify the beneficial variants, and construct new libraries based on the improved strains – all in an integrated, seamless workflow. Here we demonstrate how Iterative Genome Engineering can be used to discover novel protein variants, optimize expression, and improve heterologous protein production in yeast and E. coli strains.

Synthetic biology is disrupting diverse markets from chemicals to food, cosmetics, materials, and therapeutics. Whether you’re a metabolic engineer optimizing lysine biosynthesis in E. coli or seeking to conduct a genome-wide functional genomics screen in yeast, Digital Genome Engineering is the key to unlocking the limitless applications of synthetic biology. Realizing the full potential of this game changing technology in your research requires accessing the latest tools, understanding industry trends, and learning from best practices of industry leaders.
In partnership with SynBioBeta, Inscripta® is bringing together synthetic biology thought leaders for an innovative, FREE 2‑hour symposium. The sessions will feature perspectives from industry leaders, highlight the latest innovations in Digital Genome Engineering, and provide a vision for the future of SynBio.

Heterologous protein production is an indispensable tool in biotechnology and biopharma for manufacturing enzymes, protein therapeutics, and more. Generating robust production strains involves strategies that target both the protein expression cassette and the host genome. However, despite advances in strain engineering, optimization of protein production is still constrained by low-throughput and laborious methods that limit the success of strain optimization efforts.
In this GEN webinar, our distinguished presenter, Dr. Eric Abbate, will demonstrate a novel multiplexed genome-wide editing approach to optimize the cellobiohydrolase I enzyme (CBH1) production in S. cerevisiae. Additionally, we will learn how multiplexed and trackable editing libraries enable the discovery of new targets. These libraries comprise a wide range of edit types and targets, including genome-wide and targeted knockouts, short deletions, alternate codon, transcription factor binding sites, and terminator libraries. Finally, Dr. Abbate will describe how to identify beneficial edits across targets involved in transcription, translation, glycosylation, secretion, protein degradation, and other cellular processes, as well as evaluate the success of using targeted vs. untargeted libraries for strain engineering.

— Tycho Peterson, Managing Director, Life Science Tools & Diagnostics, J.P. Morgan
— Ipsita Smolinski, Managing Director, Capitol Street
Abstract: Join the panel to discuss the genetic technology known as synthetic biology. Two industry heavyweights — Ginkgo & Inscripta — will describe genomic engineering & organism technologies. The White House will weigh in on how it evaluates technologies and partnerships to further US science and technology priorities. One key trade association will discuss the impact synthetic biology has on non-health applications, fragrances and other flavors.

Inscripta’s Senior Director of Cell Biology, Patrick Westfall, led a panel discussion “Biological Engineering the Next Generation of Materials and Chemicals” with Kirsten Benjamin, Vice President of R&D Amyris, and Andrew Horwitz of Sestina Bio. Westfall asked the panelists about the process of selecting products they want to make and what considerations go into making those decisions.

Heterologous protein production is an indispensable tool in biotechnology and biopharma for manufacturing enzymes, protein therapeutics, and more. Generating robust production strains involves strategies that target the protein itself as well as the host genome. Despite advances in strain engineering, optimization of protein production is still limited by the ability to access the broad sequence space, constrained by traditional, low-throughput, and laborious methods like promoter substitutions or random mutagenesis. Here we use a genome-wide, multiplexed, targeted editing approach that can generate diverse edit types in both the heterologous gene and across the host genome to optimize production of cellobiohydrolase I enzyme (CHB1) in S. cerevisiae. The edits conferring improved CBH1 expression span cellular processes important for protein production, including transcription, translation, secretion, and protein degradation pathways.

Discovery in biological research was historically driven by empirical methods. The advent of biological engineering, propelled by technological developments and new tools, has made rational hypothesis-driven design and engineering of biological systems more feasible and common. Although they are often seen as opposing approaches, the empirical and rational methods can work together to accelerate fundamental research and product development. Whereas forward engineering is guided by informed design of known biological pathways and processes, the exploratory empirical methods allow to expand the design space beyond the scope of current knowledge. This webinar discusses the advantages of each method and how they could be implemented in tandem to advance biological engineering.

Heterologous protein expression in model organisms has many applications, including protein engineering, production of industrial enzymes and therapeutics, and structure-function discovery. Heterologous proteins are commonly expressed from plasmids to facilitate cloning and genetic manipulation. However, plasmid-based expression has many drawbacks, such as reduced stability and limited control over gene copy number. With CRISPR editing, chromosomal gene integration and subsequent editing has become more accessible, enabling thorough examination of protein expression and function. In this webinar, we demonstrate how we generated an

Biological responses to environmental stress can arise from multiple genetic solutions encoding tolerance mechanisms. In order to investigate the response to osmotic pressure in E. coli, we used an automated CRISPR editing workflow to generate a diverse cell population that included gene knockouts and different strength promoter substitution libraries targeting almost every gene in the genome. The resulting 25,000-variant pooled library was studied under a titrated salt challenge and the population dynamics were tracked using plasmid barcodes. By mapping 300,000 data points, we identified clear enrichment and depletion patterns and connected those back to functional genome annotations. We were able to rapidly validate our experiment with the performance of known salt stress tolerance loci, such as rpoE and osmE, and discover new gene targets.

Bacterial persistence is a potential cause of antibiotic therapy failure. Recent studies have shown that persistence is a highly evolvable trait, reaching varying frequencies within populations, depending on the selective conditions that are applied (e.g. the treatment frequency). Theory predicts that population bottlenecks, frequently encountered during host-to-host transmission and antibiotic treatment, have far-reaching effects on the evolutionary dynamics of persistence. Here, we used a combination of experimental evolution and barcoded knockout libraries to examine this hypothesis. Small bottlenecks were found to restrict the adaptive potential of populations and result in more heterogeneous evolutionary outcomes, suggest that the fitness landscape of persistence has a rugged topography. Furthermore, sequencing data revealed genes that are potentially involved in persistence, including previously known as well as novel targets.

During this webinar, discover how Inscripta experts generated an Escherichia coli saturation mutagenesis library against an integrated green fluorescent protein (GFP) using CRISPR-based technology, in a few short weeks.

The panel of industry leaders discuss technologies and microbial applications of synthetic biology in biopharma bioproduction.

In this webinar, we will discuss our work using the Onyx platform to perform genome-wide engineering of S. cerevisiae for various applications including target discovery to increase glycerol utilization, strain optimization, and forward engineering. These applications are enabled by Onyx’s ability to deliver diverse edit types that regulate gene function and expression beyond simple gene knock-outs. In total, dozens of libraries of engineered cells, containing over 100,000 precise genomic edits have been created and tested to accelerate forward engineering and genomic discovery.

Lysine, predominately used as an animal feed supplement, is a multi-billion-dollar industry however supply is limited due to the cost of current production methods. This webinar will share case studies how Inscripta leveraged its new CRISPR-based technology to design, generate, and screen libraries totaling > 200,000 single edits to improve lysine production in E. coli. Dozens of beneficial edits across the entire genome were discovered to accelerate the Design-Generate-Test-Learn cycle and increase the lysine production by 14,000-fold — ushering in the next era of genome editing.

Since its introduction, CRISPR has become a core genome editing tool that has transformed biological research. First-generation CRISPR technologies were limited in scalability, accessibility, edit variety, and ease of use, restricting their potential. Inscripta’s Onyx™ platform addresses these limitations by combining easy-to-use, intuitive software with a push-button automated benchtop device, enabling high-efficiency, massively-parallel, precision-engineered edits to Saccharomyces cerevisiae and Escherichia coli genomes. In this webinar , Tyson Shepherd will present applications in E. coli that leverage this platform. He will show how a library totaling more than 25,000 different designs and a separate library of 900 designs yielded new biochemical insights underpinning tolerance to a panel of growth-inhibitory compounds. These applications demonstrate the power of the Onyx platform to usher in a new era of genome editing.

In this webinar, we discuss our work using the Onyx platform to perform genome-wide engineering of S. cerevisiae for various applications including target discovery to increase glycerol utilization, strain optimization, and forward engineering. These applications are enabled by Onyx’s ability to deliver diverse edit types that regulate gene function and expression beyond simple gene knock-outs. In total, dozens of libraries of engineered cells, containing over 100,000 precise, genomic edits have been created and tested to accelerate forward engineering and genomic discovery.

Many of the governing rules of biology are still not known nor well understood. Thus, forward engineering of biological systems will continue to rely on empirical methods. A central challenge in forward engineering is to find genetic interventions that lead to improved function. Combinatorial optimization (fueled by large-scale diversity generation) takes ideas that show promise and recombines them in novel configurations to leverage the principles of evolutionary optimization. This optimization process is further accelerated through strategies that incorporate machine learning. Such models are interrogated to efficiently guide new library designs. Strategies and tools to effectively apply the principles of diversity generation (and combinatorial optimization) have been developed over the last two decades and are now routinely used to rapidly engineer biological systems.

Significant potential exists to harness the power of biology to better humanity and the environment, however, substantial challenges remain. Forward engineering of biological systems will continue to rely on empirical methods. Diversity generation is one of two key principles needed to reach desired outcomes in a rapid, efficient and cost-effective manner. Sequence space is vast. A central challenge in forward engineering is to find genetic interventions that lead to improved function. A large-scale diversity generation approach, where many thousands of ideas are rapidly tested, has proved to be crucial to addressing this challenge. Strategies and tools to effectively apply the principles of diversity generation (and combinatorial optimization) have been developed over the last two decades and are now routinely used to rapidly engineer biological systems.

Temperature sensitive (TS) alleles are mutations in essential genes that allow for growth at permissive temperatures and restrict or reduce growth at non-permissive temperatures. These conditional mutations are useful in academic settings for basic study of essential genes, and they are useful in bio-industrial settings as many essential genes are central to the production of high-value compounds. While critical for the study and use of essential genes, TS alleles are notoriously difficult to find. During this webinar, we will showcase how we leverage the precise, trackable genome editing of the Onyx Platform to discover novel TS alleles in yeast and E. coli. Following genome engineering, selection experiments have led to the identification of many potential TS alleles. Initial validation experiments are proving that many of these potential hits are novel TS alleles that show distinct growth properties when comparing permissive and non-permissive growth temperatures. These novel conditional mutants will function as invaluable tools for future studies and industrial applications of essential genes. This work highlights the effectiveness of Inscripta’s Digital Genome Engineering platform for novel discoveries in both yeast and E. coli.

Manufacturing of proteins, mRNA or small molecules requires comprehensive strain engineering approaches to increase yield, productivity, and robustness of the strain. While some of these goals can be accomplished with standard genetic engineering methods, others require a whole-genome, exploratory approach that can take many months. With the Onyx® Digital Genome Engineering platform, you can go after each of these objectives simultaneously by generating diverse strain libraries in a simple, automated workflow. Insertions, deletions, and substitutions across multiple targets, such as promoters, coding regions or global regulators, can be rapidly introduced to improve both specific functions and the overall manufacturability of the strain. In this webinar we will demonstrate how this approach helped improve production of heterologous proteins in E. coli and S. cerevisiae.

E. coli B‑strains, such as BL21, and their derivatives are often used for protein expression due to their optimized amino acid production, ATP utilization, lon and ompT protease deficiency, and reduced acetate accumulation. However, optimizing B‑strains for increased protein production using modern approaches to genome engineering has not been reported. Here we apply CRISPR-based, high-throughput genome-wide and pathway-specific editing to improve the expression of an enzyme. With shallow sampling of the genome-edited libraries, we identify variants with greater than 2‑fold improvement in active protein expression. This work demonstrates how rapid genome engineering can be applied to significantly enhance protein expression in E. coli.

Members of the cinnamic acid and stilbenoid chemical families are common intermediates in the production of important bioindustrial compounds. However, these molecules can inhibit microbial growth and stifle production capabilities. Since the underlying mechanism of action is unclear, ways to ameliorate inhibition can be time consuming and difficult to identify. Here we use automated, high-throughput, precise and trackable genome editing to probe biological responses to p‑coumaric acid and resveratrol. Using a combination of genome-wide and gene-specific edits, we identify genes and individual residues that show enrichment or depletion in the presence of p‑coumaric acid or resveratrol. This method represents a new approach to small molecule mechanisms-of-action discovery, providing target leads that can subsequently be used to engineer tolerance or enhance sensitivity to antimicrobial compounds.

The Design – Generate – Test – Learn (DGTL) cycle represents an efficient approach to biological engineering, from single-gene to whole-genome scale. Significant improvement can be achieved in a short amount of time by recombining beneficial edits through iterative cycles of genome editing. In order to cut down development times, all aspects of the DGLT cycle must be optimized. The Onyx™ Digital Genome Engineering Platform automates the Design and Generate steps, allowing the user to create thousands of edits, identify the beneficial variants, and construct new libraries based on the improved strains – all in an integrated, seamless workflow. Here we demonstrate how Iterative Genome Engineering can be used to discover novel protein variants, optimize expression, and improve heterologous protein production in yeast and E. coli strains.

Discovery in biological research was historically driven by empirical methods. The advent of biological engineering, propelled by technological developments and new tools, has made rational hypothesis-driven design and engineering of biological systems more feasible and common. Although they are often seen as opposing approaches, the empirical and rational methods can work together to accelerate fundamental research and product development. Whereas forward engineering is guided by informed design of known biological pathways and processes, the exploratory empirical methods allow to expand the design space beyond the scope of current knowledge. This webinar discusses the advantages of each method and how they could be implemented in tandem to advance biological engineering.

Heterologous protein expression in model organisms has many applications, including protein engineering, production of industrial enzymes and therapeutics, and structure-function discovery. Heterologous proteins are commonly expressed from plasmids to facilitate cloning and genetic manipulation. However, plasmid-based expression has many drawbacks, such as reduced stability and limited control over gene copy number. With CRISPR editing, chromosomal gene integration and subsequent editing has become more accessible, enabling thorough examination of protein expression and function. In this webinar, we demonstrate how we generated an

Biological responses to environmental stress can arise from multiple genetic solutions encoding tolerance mechanisms. In order to investigate the response to osmotic pressure in E. coli, we used an automated CRISPR editing workflow to generate a diverse cell population that included gene knockouts and different strength promoter substitution libraries targeting almost every gene in the genome. The resulting 25,000-variant pooled library was studied under a titrated salt challenge and the population dynamics were tracked using plasmid barcodes. By mapping 300,000 data points, we identified clear enrichment and depletion patterns and connected those back to functional genome annotations. We were able to rapidly validate our experiment with the performance of known salt stress tolerance loci, such as rpoE and osmE, and discover new gene targets.

During this webinar, discover how Inscripta experts generated an Escherichia coli saturation mutagenesis library against an integrated green fluorescent protein (GFP) using CRISPR-based technology, in a few short weeks.

Lysine, predominately used as an animal feed supplement, is a multi-billion-dollar industry however supply is limited due to the cost of current production methods. This webinar will share case studies how Inscripta leveraged its new CRISPR-based technology to design, generate, and screen libraries totaling > 200,000 single edits to improve lysine production in E. coli. Dozens of beneficial edits across the entire genome were discovered to accelerate the Design-Generate-Test-Learn cycle and increase the lysine production by 14,000-fold — ushering in the next era of genome editing.

Since its introduction, CRISPR has become a core genome editing tool that has transformed biological research. First-generation CRISPR technologies were limited in scalability, accessibility, edit variety, and ease of use, restricting their potential. Inscripta’s Onyx™ platform addresses these limitations by combining easy-to-use, intuitive software with a push-button automated benchtop device, enabling high-efficiency, massively-parallel, precision-engineered edits to Saccharomyces cerevisiae and Escherichia coli genomes. In this webinar , Tyson Shepherd will present applications in E. coli that leverage this platform. He will show how a library totaling more than 25,000 different designs and a separate library of 900 designs yielded new biochemical insights underpinning tolerance to a panel of growth-inhibitory compounds. These applications demonstrate the power of the Onyx platform to usher in a new era of genome editing.

In this study, we used the high-throughput CRISPR-based Onyx platform to perform saturation mutagenesis on four different essential genes involved in cell envelope synthesis in E. coli. 22,790 edits were designed. We used these saturation mutagenesis libraries to probe the essentiality of all different residues. Edits that could not be introduced suggests those residues are essential for protein function. We identified known essential amino acids (i.e. catalytic residues and residues involved in substrate binding), thereby validating our experimental approach. Several residues that were previously not known to be essential were identified. We expect our results to offer vastly improved insights into protein function, help fight against antibiotic resistance and aid structure-based drug design targeted against these essential proteins.

Temperature sensitive (TS) alleles are mutations in essential genes that allow for growth at permissive temperatures and restrict or reduce growth at non-permissive temperatures. These conditional mutations are useful in academic settings for basic study of essential genes, and they are useful in bio-industrial settings as many essential genes are central to the production of high-value compounds. While critical for the study and use of essential genes, TS alleles are notoriously difficult to find. During this webinar, we will showcase how we leverage the precise, trackable genome editing of the Onyx Platform to discover novel TS alleles in yeast and E. coli. Following genome engineering, selection experiments have led to the identification of many potential TS alleles. Initial validation experiments are proving that many of these potential hits are novel TS alleles that show distinct growth properties when comparing permissive and non-permissive growth temperatures. These novel conditional mutants will function as invaluable tools for future studies and industrial applications of essential genes. This work highlights the effectiveness of Inscripta’s Digital Genome Engineering platform for novel discoveries in both yeast and E. coli.

The Design – Generate – Test – Learn (DGTL) cycle represents an efficient approach to biological engineering, from single-gene to whole-genome scale. Significant improvement can be achieved in a short amount of time by recombining beneficial edits through iterative cycles of genome editing. In order to cut down development times, all aspects of the DGLT cycle must be optimized. The Onyx™ Digital Genome Engineering Platform automates the Design and Generate steps, allowing the user to create thousands of edits, identify the beneficial variants, and construct new libraries based on the improved strains – all in an integrated, seamless workflow. Here we demonstrate how Iterative Genome Engineering can be used to discover novel protein variants, optimize expression, and improve heterologous protein production in yeast and E. coli strains.

Heterologous protein production is an indispensable tool in biotechnology and biopharma for manufacturing enzymes, protein therapeutics, and more. Generating robust production strains involves strategies that target both the protein expression cassette and the host genome. However, despite advances in strain engineering, optimization of protein production is still constrained by low-throughput and laborious methods that limit the success of strain optimization efforts.
In this GEN webinar, our distinguished presenter, Dr. Eric Abbate, will demonstrate a novel multiplexed genome-wide editing approach to optimize the cellobiohydrolase I enzyme (CBH1) production in S. cerevisiae. Additionally, we will learn how multiplexed and trackable editing libraries enable the discovery of new targets. These libraries comprise a wide range of edit types and targets, including genome-wide and targeted knockouts, short deletions, alternate codon, transcription factor binding sites, and terminator libraries. Finally, Dr. Abbate will describe how to identify beneficial edits across targets involved in transcription, translation, glycosylation, secretion, protein degradation, and other cellular processes, as well as evaluate the success of using targeted vs. untargeted libraries for strain engineering.

Heterologous protein production is an indispensable tool in biotechnology and biopharma for manufacturing enzymes, protein therapeutics, and more. Generating robust production strains involves strategies that target the protein itself as well as the host genome. Despite advances in strain engineering, optimization of protein production is still limited by the ability to access the broad sequence space, constrained by traditional, low-throughput, and laborious methods like promoter substitutions or random mutagenesis. Here we use a genome-wide, multiplexed, targeted editing approach that can generate diverse edit types in both the heterologous gene and across the host genome to optimize production of cellobiohydrolase I enzyme (CHB1) in S. cerevisiae. The edits conferring improved CBH1 expression span cellular processes important for protein production, including transcription, translation, secretion, and protein degradation pathways.

Discovery in biological research was historically driven by empirical methods. The advent of biological engineering, propelled by technological developments and new tools, has made rational hypothesis-driven design and engineering of biological systems more feasible and common. Although they are often seen as opposing approaches, the empirical and rational methods can work together to accelerate fundamental research and product development. Whereas forward engineering is guided by informed design of known biological pathways and processes, the exploratory empirical methods allow to expand the design space beyond the scope of current knowledge. This webinar discusses the advantages of each method and how they could be implemented in tandem to advance biological engineering.

In this webinar, we will discuss our work using the Onyx platform to perform genome-wide engineering of S. cerevisiae for various applications including target discovery to increase glycerol utilization, strain optimization, and forward engineering. These applications are enabled by Onyx’s ability to deliver diverse edit types that regulate gene function and expression beyond simple gene knock-outs. In total, dozens of libraries of engineered cells, containing over 100,000 precise genomic edits have been created and tested to accelerate forward engineering and genomic discovery.

In this webinar, we discuss our work using the Onyx platform to perform genome-wide engineering of S. cerevisiae for various applications including target discovery to increase glycerol utilization, strain optimization, and forward engineering. These applications are enabled by Onyx’s ability to deliver diverse edit types that regulate gene function and expression beyond simple gene knock-outs. In total, dozens of libraries of engineered cells, containing over 100,000 precise, genomic edits have been created and tested to accelerate forward engineering and genomic discovery.

CRISPR editing has revolutionized plant breeding, and the MAD7 CRISPR nuclease developed by Inscripta<sup>TM</sup> is democratizing access to CRISPR technologies. Hudson River Biotechnology has developed a proprietary, validated molecular breeding workflow called MAD TiGER, which stands for Target identification, Guide selection, Entry into the cell and Regeneration using the MAD7 nuclease. MAD TiGER enables molecular breeding in an end-to-end fashion, providing innovative solutions to the typical barriers encountered in CRISPR plant breeding projects, such as cell entry and regeneration. The MAD TiGER workflow uses the MAD7 nuclease, nanotechnology and various synthetic gels to enable faster onset of cell divisions in regenerating and de-differentiating protoplasts towards micro-calli formation. During this webinar, Gabino Sanchez of Hudson River Biotechnology will provide an overview of the MAD TiGER workflow and discuss examples of using an integrated workflow for molecular breeding. You will also learn more about the MAD7 nuclease from Inscripta.

Bioengineering technologies hold enormous potential to solve many of our world’s pressing problems. However, they also pose biosecurity considerations. Inscripta® is committed to safe use of its genome engineering technology and has implemented multi-faceted biosecurity system designed to identify and help prevent biorisk potential of engineered organisms. Drs. Ternus, Treangen, and Vitalis will describe approaches and challenges of genome engineering biosecurity and highlight a computational tool that can predict protein function of large numbers of engineered sequence variants. The SeqScreen platform was developed by a multi-institutional team led by Signature Science and Rice University under the IARPA Fun GCAT program. Case studies will illustrate how SeqScreen serves as a comprehensive tool to report features and predicted function of novel sequences arising from bioengineering.
Documentation & Support
Inscripta® provides dedicated support through every step of your genome engineering experiments. Our Field Applications, Field Service, and Technical Support teams assist you with in-depth project consultation, customized applications design, comprehensive instrument installation, training and maintenance, and detailed data analysis and interpretation. When you are ready to begin, you can log in to the Inscripta Engineering Portal to upload genomes, design experiments, place orders, monitor instrument runs, and analyze results.
For help with our software, consumables, assays, and the Onyx® Instrument, contact us at support@inscripta.com.
Access our literature (application notes, technical notes, publications, ebooks, white papers, and MAD7 related documents) and safety data sheets for the Inscripta Onyx Platform and consumables below. User manuals, handbooks, and protocols for the Onyx platform are accessible in the Inscripta Engineering Portal for registered Onyx users.
Technology
Gene editing is the process of deliberately altering the DNA sequence of genes, from changing individual nucleotides to deleting or inserting large regions of the genome. The invention of CRISPR-based gene editing was a revolutionary breakthrough that improved the precision and efficiency of gene editing, prompting a new era of biological engineering. Learn more about the CRISPR technology and history and how Inscripta takes it to the next level.
- Copyright © 2025 Inscripta, Inc. All rights reserved.
- Legal Notices
- Privacy Policy
